Research
Unlocking the immense complexity of the cell is a major goal of modern biological science. To this end, scientists seek to acquire a detailed understanding of the many distinct molecular systems within the cell, and how these various systems function together as an integrative whole to generate unique cellular physiologies. One powerful method of obtaining such an understanding is through the identification and characterization of the protein interactions involved in these processes. In particular, large-scale proteomics studies involving the generation of complex ‘interactome’ maps of protein interconnectivity provide an unparalleled global view of the interplay between the different systems within the cell.
One major protein class of great biological importance is integral membrane proteins. Membrane proteins comprise nearly 30% of the proteome of most organisms, and have a diverse range of cellular functions, including roles as receptors, transporters, adhesion molecules, and enzymes, among others.
The association of integral membrane protein dysfunction with a range of different diseases, in addition to their accessibility and suitability for use as a target of therapeutic drugs, also makes them a protein class of significant biomedical relevance. However, the unique biochemistry and hydrophobicity of integral membrane proteins make them difficult to study using conventional biochemical approaches. Our lab is focused on the development and application of new technologies for use in the identification and characterization of the interactors of integral membrane proteins.
The Stagljar lab studies protein-protein interactions (PPIs), in particular those of membrane proteins, using new technologies that we’ve developed. Our primary interests are in the identification and characterization of the interaction partners of membrane proteins associated with disease states, to obtain a better understanding of their molecular function, and in the identification of novel drugs and drug targets for use in therapeutic treatments.
Our technologies are:
- Membrane Yeast Two-Hybrid (MYTH)
- Mammalian Membrane Two-Hybrid (MaMTH)
- Cytosolic Mammalian Two-Hybrid (CytoMath)
- Split Intein Mediated Two-Hybrid (SIMPL)
back to top
I. Membrane Yeast Two-Hybrid (MYTH)
MYTH was the original membrane PPI identification technology developed by the Stagljar lab.
Briefly, the system is based on the ‘split-ubiquitin’ principle, wherein the ubiquitin protein can be split into two stable moieties, an N-terminal fragment called Nub and a C-terminal fragment called Cub. The wild-type Nub (referred to as NubI) is capable of spontaneous reassociation with Cub to form a full-length ‘pseudo-ubiquitin’ molecule.
Mutation of isoleucine 13 to glycine in the Nub moiety (producing a fragment called NubG) prevents this spontaneous association, and allows the fragments to be adapted for use as a ‘sensor’ of protein-protein interactions. A Bait protein of interest is fused to a Cub moiety linked to a Transcription Factor (TF), while a Prey protein is fused to the NubG fragment. If the Bait and Prey do not interact, the NubG and Cub moieties remain separate. However, if an interaction between the Bait and Prey occurs, the ubiquitin moieties are brought into close proximity, allowing for pseudo-ubiquitin formation.
This pseudo-ubiquitin is recognized by cytosolic deubiquitinating enzymes (DUBs) which cleave off the transcription factor, allowing it to enter the nucleus and activate a reporter system consisting of the HIS3, ADE2 and lacZ genes. The use of appropriate selective media allows for sensitive detection of cells expressing interacting Bait-Prey pairs.
MYTH remains a powerful tool for identification of membrane protein-protein interactions, and is widely used by our collaborators and in other labs. Moreover, it was the basis for the development of additional PPI identification platforms.
back to top
II. Mammalian Membrane Two-Hybrid (MaMTH)
We developed a powerful new technology, Mammalian Membrane Two-Hybrid (MaMTH), for in vivo analysis of PPIs of full-length integral membrane proteins directly in the context of mammalian cell lines. Like MYTH, MaMTH is based upon the concept of split ubiquitin, but has been specifically re-engineered for use in the context of mammalian cells, thereby eliminating many of the complications associated with studying mammalian proteins in a foreign host.
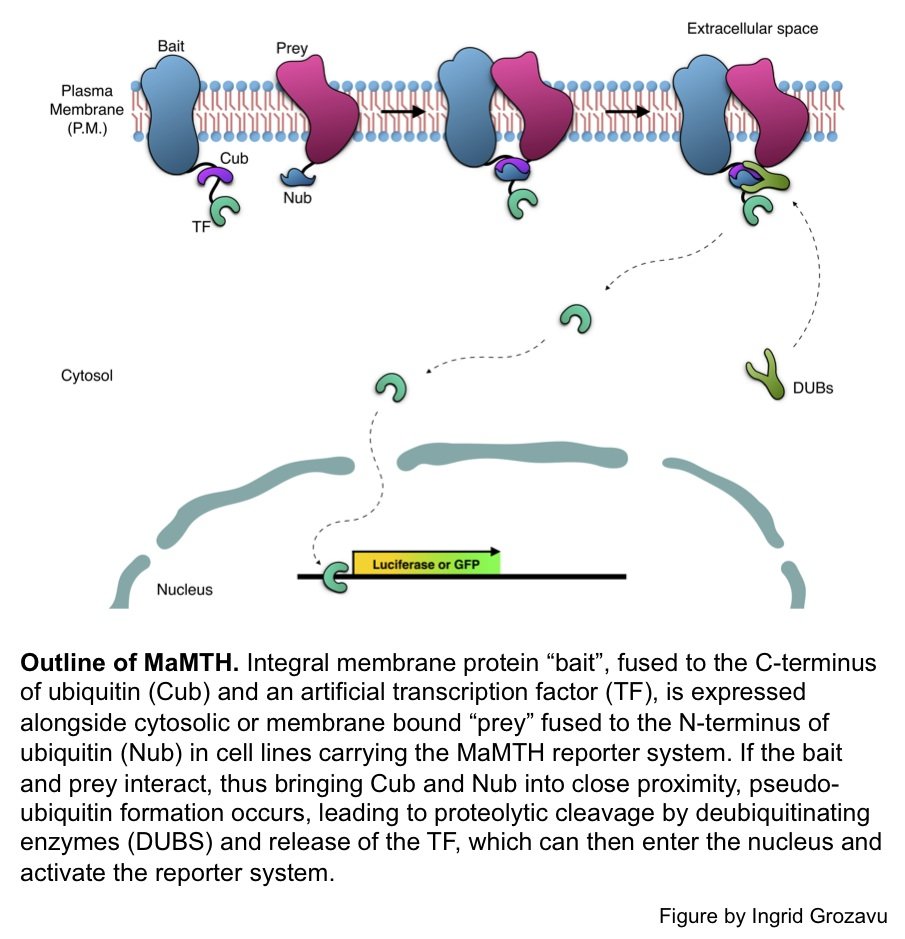
A major advantage of this system is its sensitivity, which allows it to detect subtle dynamic changes in interaction patterns in response to environmental changes (e.g. drugs, ligands, phosphorylation state changes etc.). Additionally the system is highly transferable, allowing it to be carried out in virtually any cell line, and is perfectly suited for use in a high-throughput format.
Using MaMTH as a targeted protein interaction screening assay, we identified a protein called CRKII as a novel interactor of oncogenic EGFR (L858R) and showed that CRKII promotes persistent activation of aberrant signaling in non-small cell lung cancer (NSCLC) cells, thus identifying CRK II as a novel potential biomarker for NSCLC (Petschnigg et al. (2014) Nature Methods). To the best of our knowledge, high-throughput dynamic interaction screens (i.e. performed under multiple conditions for comparison) with full-length membrane proteins in human cells have not been possible in the field of proteomics thus far.
Current MaMTH projects include:
- Disease-associated PPIs
- MaMTH drug-screening platform
- High-throughput MaMTH
back to top
Disease-associated PPIs
Membrane PPIs are associated with numerous diseases, including cancers, cystic fibrosis and neurological disorders such as Alzheimer's disease and Parkinson's disease. By mapping the interactomes of disease-associated PPIs, we can learn more about the underlying mechanisms and identify potential therapeutic targets. Some ongoing MaMTH projects are described below.
RTK-SH2/PTB interactome
Receptor Tyrosine Kinases (RTKs) are an important family of membrane proteins that are implicated in cancers. RTKs bind extracellular ligands and convert that biological event into a signal to the cell, through dimerization and auto phosphorylation. This initiates signalling cascades of protein-protein interactions, which result in specific gene expression to the signal. Downstream signals convey information required for constant expression of genes for growth, transformation, movement, and proliferation.
Using MaMTH technology, we have already screened multiple full length RTK baits with all human SH2- and PTB-domain containing preys and identified novel hits, of which we have validated some functionally, with dramatic cancer viability consequences.
MET
Aberrant signalling of the MET tyrosine kinase receptor underlies tumour phenotypes such as hyper-proliferation, metastasis and cancer drug resistance. MET is found to aggravate cancers rather than drive them, hence is a prime target in combinatorial cancer therapies. However, biological information is scarce. By understanding the protein-protein interactome of MET, we can uncover novel functions, regulatory mechanisms and therapeutic targets. The collective aim of the MET interactome project is to map both targeted and global protein-protein interactomes using MaMTH technology and follow-up by elucidating the functional significance of novel interactors. At the culmination of this project, we hope to provide a more comprehensive view of MET interactome and by extension its physiological and pathophysiological function, regulatory partners and potential targets for MET targeting therapies.
ALK
Anaplastic Lymphoma Kinase (ALK), which belongs to the RTK family, plays a critical role in development and progression of many cancers. To design new treatments for ALK-associated diseases, it is essential to study the interactions of the protein to better understand the biological significance of its protein-protein interactions (PPIs) and how they contribute to ALK regulation and function. MaMTH was used in this study to generate a targeted protein-protein interaction map of full length ALK. The final, overall aim of future work will be a global, unbiased screen of the entire human protein-encoding open reading frame collection and functional characterization of novel PPIs of ALK to identify disease-associated interactors.
K-RAS
Kirsten Rat Sarcoma (K-RAS) is the first described human oncogene, dating back to 1982, and is found in up to 30% of all human cancers. While much of the underlying biology has already been elucidated, oncogenic K-RAS isoforms remain clinically undruggable. However, using MaMTH-HTS, our lab plans to build a truly comprehensive and dynamic interaction of K-RAS (i.e. in WT and oncogenic form). This map in combination with MaMTH-DS will help uncover K-RAS-targeting drugs, and predict better combinational therapy for mutant K-RAS-harbouring cancers.
CFTR
Mutations in Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) gene cause Cystic Fibrosis, a disease that leads to progressive loss of respiratory function. We are studying the dynamic interactome of CFTR protein, which takes into account the change in protein-protein interactions (PPIs) that can occur in the presence of drugs, and between mutant and wild-type forms of CFTR. Using MaMTH, we are identifying novel interacting protein partners of both wildtype and mutant CFTR and investigating the mechanisms of protein interactions to develop unique CF therapy discovery platforms. This will provide insight into the molecular machinery affecting CFTR in CF and will be of great value in the development of future therapeutics. Membrane potential assays are performed on the interacting partner candidates as follow-up functional studies along with a pilot drug screening study. Our strategy represents the first application of MaMTH technology to identify small molecule interaction modulators effective in correcting CFTR trafficking and has far-reaching application to other membrane trafficking diseases.
back to top
MaMTH-DS (Drug Screening)
The MaMTH approach enables quantitative measurement of dynamic protein-protein interactions (PPIs) in vivo in the natural membrane environment of human cells. The system addresses a currently challenging area of transmembrane protein analysis, which is of keen interest to the pharmaceutical market, as membrane proteins are a major class of drug targets.
Recently, we have adapted MaMTH into a high-throughput, small-molecule screening platform, called MaMTH-DS, to identify compounds specifically targeting functional interactions of receptor tyrosine kinases (RTKs). In this assay, an inhibiting compound that affects RTK kinase activity will result in reduction of the phosphorylation status of a given RTK, leading to reduction in binding of an adapter 'prey' protein such as Shc1 or Grb2. As proof-of principle, we used MaMTH-DS to screen a pilot collection of 2,960 small molecules against cells expressing the EGFR L858R/T790M/C797S mutant (associated with drug-resistant NSCLC) and identified three novel compounds specifically targeting the functionally important interaction of this triple mutant protein with the Shc1 adapter protein. One of these compounds will soon be the subject of a Phase II drug repurposing clinical trial being conducted by the Thoracic Oncology Program at the Princess Margaret Cancer Centre, Toronto.
We believe that our technology will provide a means of both increasing our basic knowledge and promoting drug-discovery, thereby making significant contributions to human health care and research.
back to top
MaMTH-HTS (High-throughput MaMTH)
We are currently working to expand our MaMTH assay into platforms suitable for the high-throughput screening of complex libraries (MaMTH-HTS).
- Pooled ORFeome: In this approach, the luciferase reporter used in our traditional MaMTH assay is replaced with the eGFP protein, allowing for the selective isolation of cells containing interactions from mixed populations using fluorescence-activated cell sorting (FACS).
- Array MaMTH: preys are introduced to stably-expressed baits in luciferase reporter cells in an arrayed format. Interactors are easily identified by a luciferase signal at their location in the array.
We will use these MaMTH-HTS platforms to probe PPIs of proteins associated with neurological diseases and cancers. Additionally, in collaboration with the laboratory of Professor T. Rabbitts at the Weatherall Institute of Molecular Medicine, University of Oxford, we are using pooled ORFeome MaMTH-HTS to screen complex libraries of intracellular single domain antibodies (iDabs) in an effort to find strong binders of both oncogenic KRAS and HRAS proteins, which can be used as therapeutics to directly disrupt functional interactions critical to disease.
back to top
III. Cytosolic Mammalian Two-Hybrid (CytoMaTH)
Membrane proteins behave in association with downstream signaling effectors. Thus, a study of these proteins is incomplete without accounting for their effector pathways. For this reason, we are currently engineering a MaMTH-derived assay to probe for cytosolic PPIs.
To enable this, we introduced several important modifications to our MaMTH bait construct, which allowed for both a decrease in the background signal produced solely by cytosolic baits, but also maintained the assay's integrity at detecting PPIs, both cytosolic and in the plasma membrane.
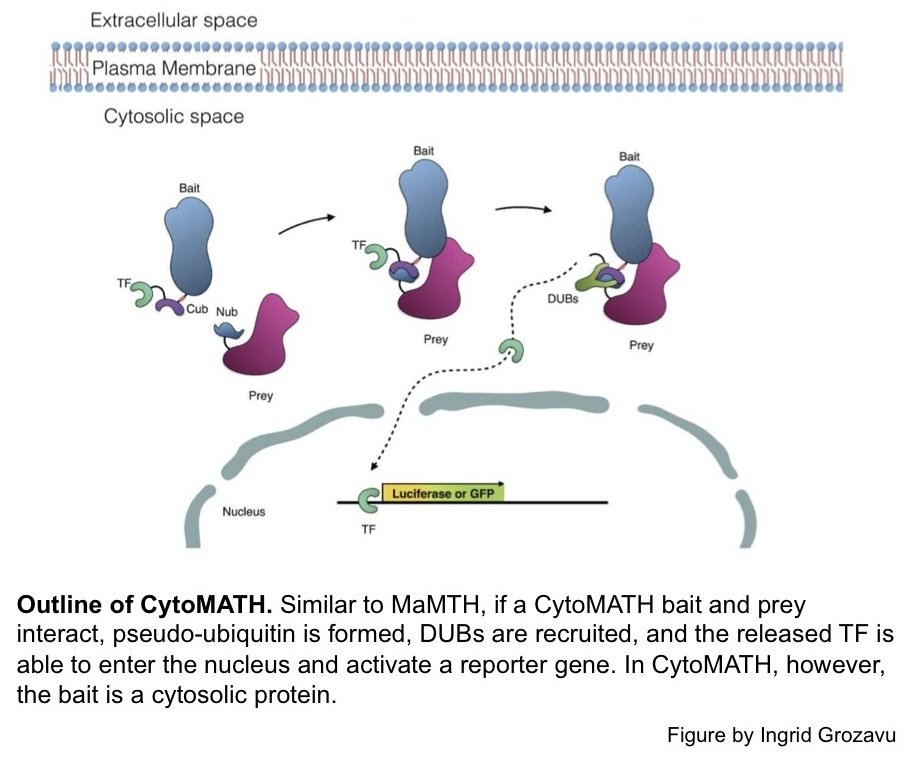
CytoMaTH is currently being tested against an array of bait and prey proteins.
back to top
IV. Split-Intein Mediated Protein Ligation (SIMPL)
Split Intein-Mediated Protein Ligation (SIMPL) is a new technology developed in our lab that can be applied to any cellular space, either within or outside the cell. Split inteins are proteins that occur naturally in all cells. They are in two parts, each part attached to another peptide (exteins), and when the two split intein parts come together they cause the the exteins to fuse with each other, a process called protein splicing. To harness this as a PPI detection tool, SIMPL, we engineered the split intein to abolish self-association. The split intein parts can be attached to two proteins, and splicing occurs if they interact.
Preliminary results demonstrate that SIMPL works in real-time and can detect short-lived, conditional and weak interactions in addition to stable and strong PPIs. We are currently developing SIMPL into a method for rapid identification of genome-wide interactors of any given protein. We also anticipate its use as a drug screening platform to search for drug-like compounds that are able to prevent harmful protein-protein interactions from forming.
back to top