
Christopher Ing
I aim to understand the biophysical mechanisms of wild-type and mutant cytochrome c oxidase using all-atom molecular dynamics. I am particularly interested in ion movement and solvation in these proteins.
Research Interests
Under the guidance of Dr. Pierre Nicholas Roy, I calculated properties of small quantum clusters systems at ultra low temperatures. To achieve this I utilized a path integral molecular dynamics sampling method which I assisted in implementing in the Molecular Modelling Toolkit. The simulation of path integral helium about a fixed CO2 impurity is shown here for a 64 time-slice helium atom.
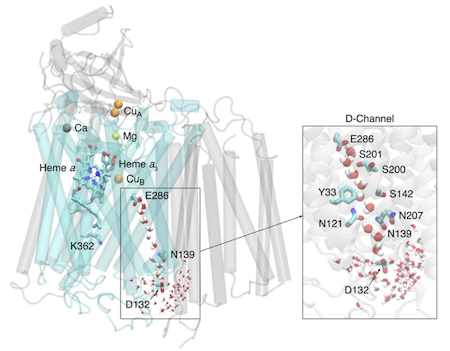
In my current research, I aim to advance research in the Pomès Lab on the molecular structure and function of cytochrome c oxidase. Cytochrome c oxidase of R. sphaeroides is depicted in the following figure from J. Phys.: Condens. Matter 23 (2011) 234102. Subunit I, which contains the D channel (inset) and the binuclear centre, highlighted in blue. These are the primary regions of importance to electron and proton transport.
I am also actively involved in studying the molecular structure and function of other membrane proteins like the voltage-gated sodium channel. I am currently working on Python software to assist in the analysis of timeseries data from multiple simulation repeats.

