McQuibban Lab

Department of Biochemistry, University of Toronto



Our Research
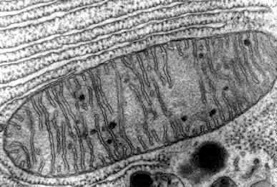
Since the 1940s, the mitochondrion has been known to be the powerhouse of the cell, due to its primary role in generating most cellular energy in the form of ATP. Mitochondria have been depicted as static, kidney bean shaped organelles dispersed throughout the cytoplasm.
An electron micrograph depicting the classic view of mitochondria
However, it is now clear that mitochondria are dynamic organelles, consisting of interconnected tubular networks constantly undergoing rapid directed movements along microtubules combined with multiple and balanced membrane fusion/fission reactions, even in resting cells. The reason for this dynamic behaviour of mitochondria is just beginning to be understood. Disruption of mitochondrial dynamics is associated with neurodegenerative diseases such as dominant optic atrophy, Charcot-Marie-Tooth type 2A and Parkinson’s disease. Previous studies have identified only a handful of factors that are involved in regulating mitochondrial membrane dynamics.
Fluorescence microscopy movie showing dynamic mitochondria undergoing fusion and fission in a Drosophila S2 cell
My lab is interested in determining the protein machinery that both orchestrates and regulates mitochondrial membrane dynamics, particularly fusion.
We also aim to understand, on a broader scale, the biological significance of dynamic mitochondria, and the consequences of disrupting this process in relation to human mitochondrial diseases. To do this, we are taking advantage of the power of yeast genetics to specifically identify factors that control membrane fusion. We are also using the fruit fly, Drosophila melanogaster, as a model metazoan, to dissect in what cell-biological context a dynamic mitochondrial compartment is required, an area of research that has yet to be pursued in this model organism. Additionally, we use molecular and cell biological techniques in mammalian tissue culture to explore the basic functional mechanisms of proteins involved in mitochondrial dynamics.
The purpose of our research is to investigate two key questions in this emerging field:
What role do mitochondrial dynamics play in cell and tissue function?
What molecular machinery is required to orchestrate and regulate mitochondrial double membrane fusion?
To address these questions we are currently undertaking the following four specific aims:
1. Identify and characterize genes required specifically for mitochondrial membrane fusion in yeast.
We are using the yeast, Saccharomyces cerevisiae, to identify novel factors that orchestrate and regulate mitochondrial membrane fusion. By taking advantage of the existing yeast deletion collection, we are both candidate testing, and adopting a high-throughput approach. Genes that are identified in these screens will be further characterized in yeast, and in flies (assuming they are conserved through evolution). Early results from these screens are leading us to explore the interplay between ER and mitochondrial cross-talk pathways including phospholipid exchange and ubiquitin-mediated protein degradation.
Fluorescence microscopy image of a living yeast cell expressing a mitochondrial targeted GFP reveals a tubular, interconnected network at the cell cortex.
2. Determine the substrate repertoire, in mammals and in flies, of the mitochondrial rhomboid, a protease that is an upstream regulator of mitochondrial dynamics.
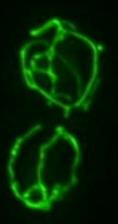
Our introduction to mitochondrial membrane dynamics came from our discovery that a mitochondria-localized rhomboid protease regulated this process. Rhomboids are 7-pass transmembrane (TM) domain proteins that cleave their substrates in the lipid bilayer using a classical serine catalytic-triad mechanism (like trypsin) and represent a new conserved family of intramembrane proteases. In flies, we have identified two substrates for the mitochondrial protease, Pink1 and Omi/Htra2. These two proteins are key regulators of cellular viability and mutations in in the genes for Pink1 and Omi/Htra2 are associated with Parkinson’s disease. More recently, we have shown that the mitochondrial rhomboid cleaves Pink1 in humans and have found a mutation in the human mitochondrial rhomboid gene, known as PARL, that is associated with Parkinson’s disease. Pink1 has emerged recently as an important regulator of mitophagy, the regulated destruction of damaged mitochondria. Our work is now focussed on determining how PARL cleavage of Pink1 regulates this mitochondrial quality control pathway.
Fluorescence microscopy image of a Drosophila S2 cell showing the interconnected mitochondrial network.
3. Using structural and biochemical techniques, determine the biophysical mechanism of Mgm1-mediated inner membrane fusion.
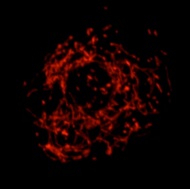
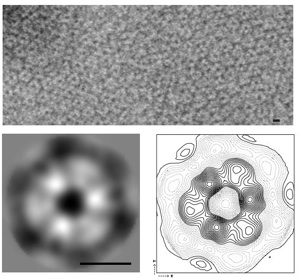
In yeast, we have discovered that the main action of the mitochondrial rhomboid is to regulate the cleavage of Mgm1. Mgm1 is a master regulator of membrane fusion whose mammalian ortholog, OPA1, is associated with optic atrophy type I, a genetic optic neuropathy disorder that causes blindness. Mgm1 is a dynamin-like GTPase that coordinates the fusion of the inner mitochondrial membrane through an interaction between a long, membrane tethered form and a soluble, short, rhomboid-cleaved form of the protein. We have determined that Mgm1’s GTPase activity is stimulated by binding to phosopholipids and that it can assemble into an ordered lattice on liposomes. Future studies will focus on determining how the long form and short form work together to orchestrate membrane fusion.
Electron micrograph showing purified Mgm1 protein assembles into oligomeric rings on liposomal membranes
4. Fully characterize in the model organism, Drosophila melanogaster, the tissue-specific requirements for mitochondrial dynamics using genetics and mutant fly analyses.
We have made a fly that has a mutation in its mitochondrial rhomboid (rhomboid-7). These flies are semi-viable. Homozygous mutant adults only live for three days (normal flies live for about 60 days). In addition, these flies cannot walk or fly, the consequence of a severe neuromuscular defect. We are currently characterizing the phenotype of this mutant, by a number of genetic and biochemical approaches. It is our hope that by studying mitochondrial function in a complex metazoan, we will be able to understand better in what cellular context mitochondrial membrane dynamics are required for function.
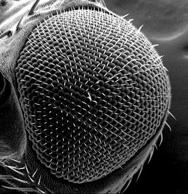
Scanning EM of the Drosophila compound eye showing the crystalline ommatidial array. Its sensitivity to disruption is the basis of many genetic screens.
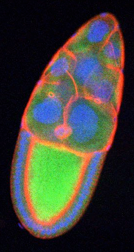
Fluorescence microscopy image of a Drosophila ovary showing mitochondria (green), cytoskeleton (red) and nuclei (blue).