 |
 |
 |
 |
  |
Hue Sun
CHAN
Professor
B.Sc., University
of Hong Kong, 1981
M.A., University
of California at Berkeley, 1983
Ph.D., University
of California at Berkeley, 1987
Postdoctoral Fellow,
University of California at San
Francisco (UCSF), 1987-89
Assistant Research Biophysicist,
Assistant Adjunct Professor,
Associate Adjunct Professor,
UCSF, 1989-98
Associate Professor,
University of Toronto,
1998-2003
Canada Research
Chair, 2001-2010
Professor,
University of Toronto,
2003-present
Teaching:
BCH340H -- Proteins: From Structure
to Proteomics (Archive)
JBB2026H -- Protein Structure, Folding and Design (offered NOW:
Current Fall '14 course outline)
Member of:
CIHR Training Program (at U. of Toronto)
in Protein Folding and Interaction Dynamics:
Principles & Diseases
(Program Advisory Committee)
Editorial Board:
Proteins: Structure, Function, and Bioinformatics
|
Medical
Sciences Building, Room 5363
416-978-2697
chan@arrhenius.med.utoronto.ca |
|

Theoretical and Computational Investigations of Protein Folding, Interactions, and Evolution



Research Synopsis
 |
|

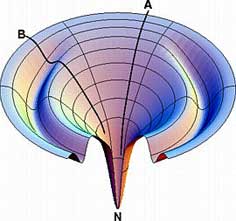
 Moat Landscape:
a protein could have a fast-folding
throughway process (A) in parallel with a slow-folding
process (B) involving a kinetic trap.
Moat Landscape:
a protein could have a fast-folding
throughway process (A) in parallel with a slow-folding
process (B) involving a kinetic trap.
From Levinthal to pathways to funnels,
Nature Structural Biology, Volume 4, No. 1,
January 1997.
Current Research Group Members
Research
Group Pictures
U. of
Toronto pictures
Biophysics in Canada:
Links to
other Canadian biophysics groups
(kindly provided by Andrew Rutenberg)
|
Please see
H.S. Chan's
departmental page for a
more detailed summary & graphical illustrations of his research group's
current interests and projects.
Protein folding and interactions are physico-chemical processes. Our
group's overall research goal is to elucidate their underlying
energetics. To this end, a main emphasis of our effort is to develop
proteinlike heteropolymer models with coarse-grained interactions and
simplified representations of chain geometries. The rationale of these
approaches is to capture the essential physics and at the same time
allow for a broad coverage of the protein conformational space -- and
also a broad coverage of the sequence space for evolutionary studies --
that is not readily achievable currently in higher-resolution models.
Molecular dynamics simulations using common atomic forcefields and
explicit water models are used in our work as well, especially for
deciphering subtle properties of solvent-mediated interactions. Various
combinations of coarse-grained and atomic methods are being used to
gain physical insights into general principles of folding, protein
interactions, and evolution. The topics we address including folding
cooperativity, origin of enthalpic and volumetric folding barriers,
nonnative effects in folding, formation of functional and disease-causing
dynamic, "fuzzy" complexes involving intrinsically disordered proteins,
and conformational switching in protein evolution. Some of these efforts,
including investigations into the mathematical basis of the unknotting,
decatenating, and supercoil simplifying actions of type-2 topoisomerases,
are highlighted in
H.S. Chan's departmental webpage
(please click here).

|
|


Selected Publications
 |
Biophysics of Protein Evolution and Evolutionary Protein Biophysics.
T. Sikosek & H.S. Chan
J. Royal Soc. Interface 11:20140419 (2014).
Polycation-π Interactions are a Driving Force for Molecular
Recognition by an Intrinsically Disordered Oncoprotein Family.
J. Song, S. C. Ng, P. Tompa, K. A. W. Lee & H. S. Chan,
PLoS Comput. Biol. 9(9):e1003239 (2013).
Transition Paths, Diffusive Processes, and Preequilibria of Protein Folding.
Z. Zhang & H. S. Chan, Proc. Natl. Acad. Sci.
USA 109:20919-20924 (2012).
Evolutionary Dynamics on Protein Bi-Stability Landscapes can Potentially
Resolve Adaptive Conflicts.
T. Sikosek, E. Bornberg-Bauer & H. S. Chan, PLoS Comput. Biol.
8(9):e1002659 (2012).
Escape from Adaptive Conflict Follows from Weak Functional Trade-Offs
and Mutational Robustness.
T. Sikosek, H. S. Chan & E. Bornberg-Bauer, Proc. Natl. Acad. Sci.
USA 109:14888-14893 (2012).
Cooperativity, Local-Nonlocal Coupling, and Nonnative Interactions:
Principles of Protein Folding from Coarse-Grained Models.
H. S. Chan, Z. Zhang, S. Wallin & Z. Liu, Annu. Rev. Phys. Chem.
62:301-326 (2011).
Action at Hooked or Twisted-Hooked DNA Juxtapositions Rationalizes
Unlinking Preference of Type-2 Topoisomerases. Z. Liu, L. Zechiedrich
& H. S. Chan, J. Mol. Biol. 400:963-982 (2010).
Competition Between Native Topology and Nonnative Interactions in
Simple and Complex Folding Kinetics of Natural and Designed Proteins.
Z. Zhang & H. S. Chan,
Proc. Natl. Acad. Sci. USA 107:2920-2925 (2010).
Desolvation Barrier Effects are a Likely Contributor to the Remarkable
Diversity in the Folding Rates of Small Proteins.
A. Ferguson, Z. Liu & H. S. Chan,
J. Mol. Biol. 389:619-636 (2009).
Theoretical and Experimental Demonstration of the Importance of Specific
Nonnative Interactions in Protein Folding.
A. Zarrine-Afsar, S. Wallin, A. M. Neculai, P. Neudecker, P. L. Howell,
A. R. Davidson & H. S. Chan, Proc. Natl. Acad. Sci. USA
105:9999-10004 (2008).
Polyelectrostatic Interactions of Disordered Ligands Suggest a Physical
Basis for Ultrasensitivity.
M. Borg, T. Mittag, T. Pawson, M. Tyers, J. D. Forman-Kay & H. S. Chan,
Proc. Natl. Acad. Sci. USA 104:9650-9655 (2007).
Hydrophobic Association of α-Helices, Steric Dewetting and Enthalpic
Barriers to Protein Folding.
J. L. MacCallum, M. Sabaye Moghaddam, H. S. Chan & D. P. Tieleman,
Proc. Natl. Acad. Sci. USA 104:6206-6210 (2007).
Topological Information Embodied in Local Juxtaposition Geometry
Provides a Statistical Mechanical Basis for Unknotting by Type-2
DNA Topoisomerases.
Z. Liu, J. K. Mann, E. L. Zechiedrich & H. S. Chan,
J. Mol. Biol. 361:268-285 (2006).
Desolvation is a Likely Origin of Robust
Enthalpic Barriers to Protein Folding.
Z. Liu & H. S. Chan, J. Mol. Biol. 349:872-889 (2005).
Temperature Dependence of Three-Body Hydrophobic Interactions: Potential
of Mean Force, Enthalpy, Entropy, Heat Capacity, and Nonadditivity.
M. Sabaye Moghaddam, S. Shimizu & H. S. Chan,
J. Am. Chem. Soc. 127:303-316 (2005).
Sparsely Populated Folding Intermediates of the Fyn SH3 Domain:
Matching Native-Centric Essential Dynamics and Experiment.
J. E. Ollerenshaw, H. Kaya, H. S. Chan & L. E. Kay,
Proc. Natl. Acad. Sci. USA 101:14748-14753 (2004).
Cooperativity Principles in Protein Folding.
H. S. Chan, S. Shimizu & H. Kaya, Methods
Enzymol. 380:350-379 (2004).
Origins of Chevron Rollovers in Non-Two-State Protein Folding Kinetics.
H. Kaya & H. S. Chan, Phys. Rev. Lett. 90:258104 (2003).
Recombinatoric Exploration of Novel Folded Structures:
A Heteropolymer-Based Model of Protein Evolutionary
Landscapes. Y. Cui, W. H. Wong, E. Bornberg-Bauer &
H. S. Chan, Proc. Natl Acad. Sci. USA 99:809-814 (2002).
Conformational Propagation with Prion-like
Characteristics in a Simple Model of Protein
Folding. P. M. Harrison, H. S. Chan, S. B. Prusiner &
F. E. Cohen, Protein Sci. 10:819-835 (2001).
Folding Alphabets. H. S. Chan, Nature Struct. Biol. 6:994-996
(1999).
Energetic Components of Cooperative Protein Folding.
H. Kaya & H. S. Chan, Phys. Rev. Lett. 85:4823-4826 (2010).
Modeling Evolutionary Landscapes: Mutational Stability, Topology and
Superfunnels in Sequence Space.
E. Bornberg-Bauer & H. S. Chan, Proc. Natl. Acad. Sci. USA
96:10689-10694 (1999).
Protein Folding: Matching Speed and Locality.
H. S. Chan, Nature 392:761-763 (1998).
Protein
Folding in the Landscape Perspective: Chevron Plots
and Non-Arrhenius Kinetics. H. S. Chan & K. A. Dill, Proteins:
Struct. Funct. Genet. 30:2-33 (1998).
From
Levinthal to Pathways to Funnels. K. A. Dill & H. S. Chan,
Nature Struct. Biol. 4:10-19 (1997). |
|
Click
here for an extended list of publications
and electronic reprints.
Lectures
and seminars
Journal
cover images
Meetings
organized |
|
 |





















